dag_paths finds open paths between a given exposure and outcome.
ggdag_paths and ggdag_paths_fan plot all open paths. See
dagitty::paths() for details.
Usage
dag_paths(
.dag,
from = NULL,
to = NULL,
adjust_for = NULL,
limit = 100,
directed = FALSE,
paths_only = FALSE,
...
)
ggdag_paths(
.tdy_dag,
from = NULL,
to = NULL,
adjust_for = NULL,
limit = 100,
directed = FALSE,
shadow = TRUE,
...,
size = 1,
edge_type = c("link_arc", "link", "arc", "diagonal"),
node_size = 16,
text_size = 3.88,
label_size = text_size,
text_col = "white",
label_col = "black",
edge_width = 0.6,
edge_cap = 8,
arrow_length = 5,
use_edges = TRUE,
use_nodes = TRUE,
use_stylized = FALSE,
use_text = TRUE,
use_labels = FALSE,
label_geom = geom_dag_label_repel,
text = NULL,
label = NULL,
node = deprecated(),
stylized = deprecated()
)
ggdag_paths_fan(
.tdy_dag,
from = NULL,
to = NULL,
adjust_for = NULL,
limit = 100,
directed = FALSE,
...,
shadow = TRUE,
spread = 0.7,
size = 1,
node_size = 16,
text_size = 3.88,
label_size = text_size,
text_col = "white",
label_col = "black",
edge_width = 0.6,
edge_cap = 8,
arrow_length = 5,
use_edges = TRUE,
use_nodes = TRUE,
use_stylized = FALSE,
use_text = TRUE,
use_labels = FALSE,
label_geom = geom_dag_label_repel,
unified_legend = TRUE,
text = NULL,
label = NULL,
node = deprecated(),
stylized = deprecated()
)Arguments
- .dag
A
tidy_dagittyordagittyobject- from
A character vector with starting node name(s), or
NULL. IfNULL, checks DAG for exposure variable.- to
A character vector with ending node name(s), or
NULL. IfNULL, checks DAG for outcome variable.- adjust_for
character vector, a set of variables to control for. Default is
NULL.- limit
maximum amount of paths to show. In general, the number of paths grows exponentially with the number of variables in the graph, such that path inspection is not useful except for the most simple models.
- directed
logical. Should only directed paths be shown?
- paths_only
logical. Should only open paths be returned? Default is
FALSE, which includes every variable and edge in the DAG regardless if they are part of the path.- ...
additional arguments passed to
tidy_dagitty()- .tdy_dag
A
tidy_dagittyordagittyobject- shadow
logical. Show edges which are not on an open path?
- size
A numeric value scaling the size of all elements in the DAG. This allows you to change the scale of the DAG without changing the proportions.
- edge_type
The type of edge, one of "link_arc", "link", "arc", "diagonal".
- node_size
The size of the nodes.
- text_size
The size of the text.
- label_size
The size of the labels.
- text_col
The color of the text.
- label_col
The color of the labels.
- edge_width
The width of the edges.
- edge_cap
The size of edge caps (the distance between the arrowheads and the node borders).
- arrow_length
The length of arrows on edges.
- use_edges
A logical value. Include a
geom_dag_edges*()function? IfTRUE, which is determined byedge_type.- use_nodes
A logical value. Include
geom_dag_point()?- use_stylized
A logical value. Include
geom_dag_node()?- use_text
A logical value. Include
geom_dag_text()?- use_labels
A logical value. Include a label geom? The specific geom used is controlled by
label_geom.- label_geom
A geom function to use for drawing labels when
use_labels = TRUE. Default isgeom_dag_label_repel. Other options includegeom_dag_label,geom_dag_text_repel,geom_dag_label_repel2, andgeom_dag_text_repel2.- text
The bare name of a column to use for
geom_dag_text(). Ifuse_text = TRUE, the default is to usename.- label
The bare name of a column to use for labels. If
use_labels = TRUE, the default is to uselabel.- node
Deprecated.
- stylized
Deprecated.
- spread
the width of the fan spread
- unified_legend
A logical value. When
TRUEand bothuse_edgesanduse_nodesareTRUE, creates a unified legend entry showing both nodes and edges in a single key, and hides the separate edge legend. This creates cleaner, more compact legends. Default isTRUE.
Value
a tidy_dagitty with a path column for path variables, a set
grouping column, and a path_type column classifying paths as "backdoor"
or "direct", or a ggplot.
Examples
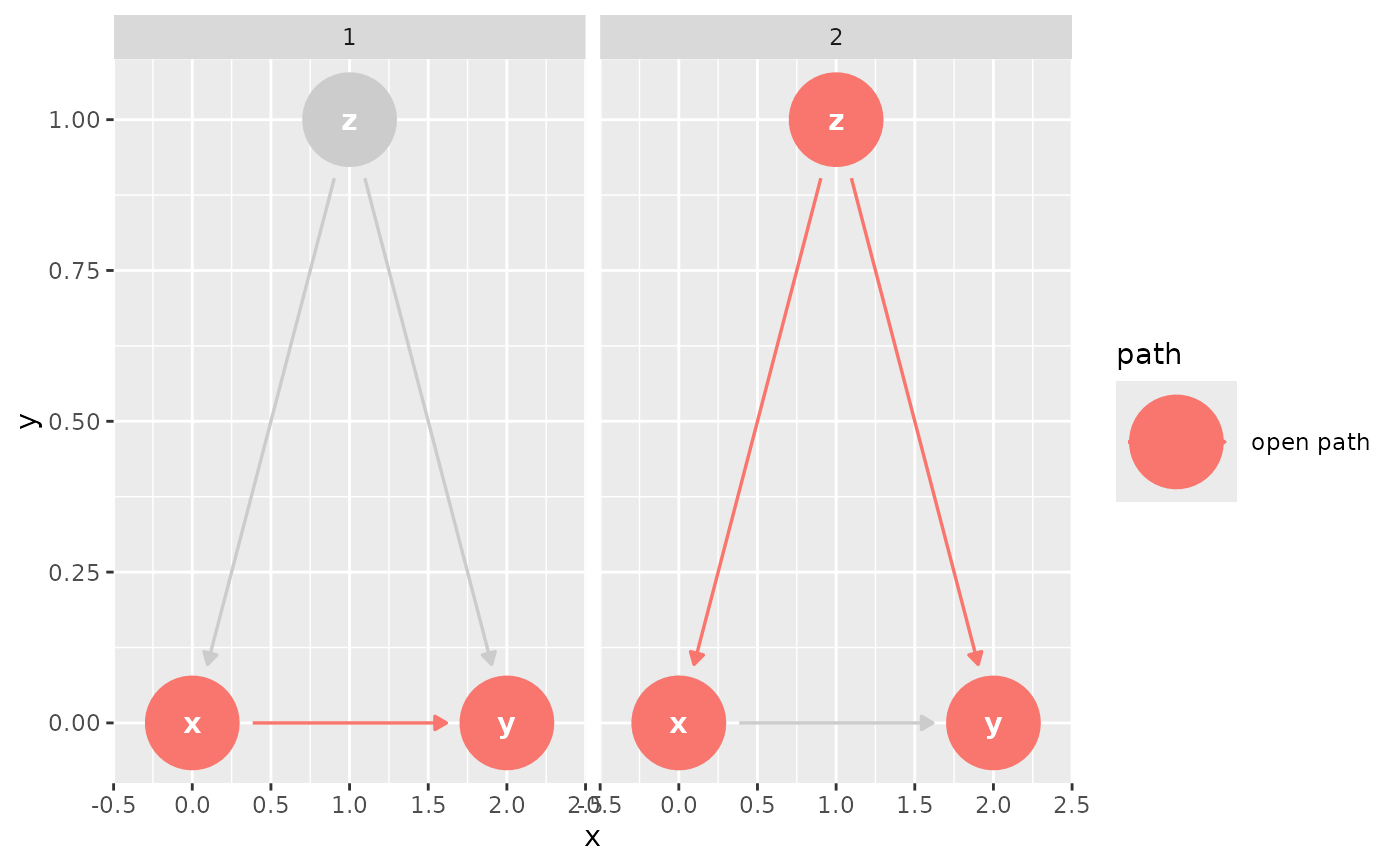
confounder_triangle(x_y_associated = TRUE) |>
dag_paths(from = "x", to = "y")
#> # DAG:
#> # A `dagitty` DAG with: 3 nodes and 3 edges
#> # Exposure: x
#> # Outcome: y
#> # Paths: 2 open paths: {x -> y}, {x <- z -> y}
#> #
#> # Data:
#> # A tibble: 9 × 10
#> set name x y direction to xend yend path path_type
#> <chr> <chr> <int> <int> <fct> <chr> <int> <int> <chr> <chr>
#> 1 1 x 0 0 -> y 2 0 open path direct
#> 2 1 y 2 0 NA NA NA NA open path direct
#> 3 1 z 1 1 -> x 0 0 NA NA
#> 4 1 z 1 1 -> y 2 0 NA NA
#> 5 2 x 0 0 -> y 2 0 NA NA
#> 6 2 y 2 0 NA NA NA NA open path backdoor
#> 7 2 z 1 1 -> x 0 0 open path backdoor
#> 8 2 z 1 1 -> y 2 0 open path backdoor
#> 9 2 x 0 0 NA NA NA NA open path backdoor
#> #
#> # ℹ Use `pull_dag() (`?pull_dag`)` to retrieve the DAG object and `pull_dag_data() (`?pull_dag_data`)` for the data frame
confounder_triangle(x_y_associated = TRUE) |>
ggdag_paths(from = "x", to = "y")
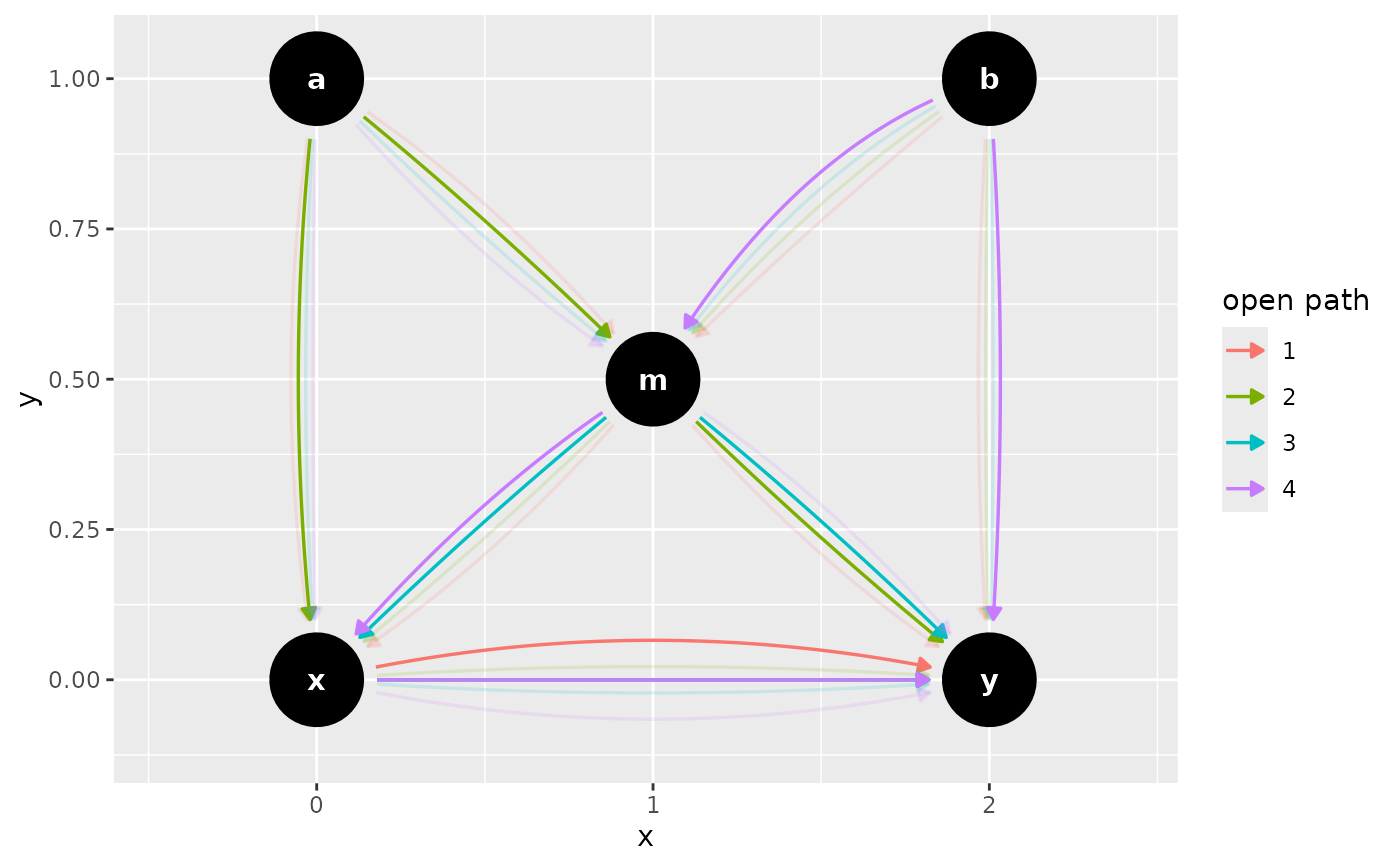
 butterfly_bias(x_y_associated = TRUE) |>
ggdag_paths_fan(shadow = TRUE)
butterfly_bias(x_y_associated = TRUE) |>
ggdag_paths_fan(shadow = TRUE)